Merged Peptides View Page¶
To reach this page, select multiple searches on the project page and click “View Merged Peptides”. (See Project Overview Page.) This page combines and collates the data from multiple searches and presents the results as an interactive table. The searches do not need to be from the same software pipeline. For example, different versions of the same program may be compared, or the results from entirely different programs (e.g., Kojak and XQuest) may be compared. Currently, the total number of merged searches must be 2 or 3 and must be from the same project. Note, for the peptide view page seen when viewing a single search, see Peptides View Page.
Search List¶
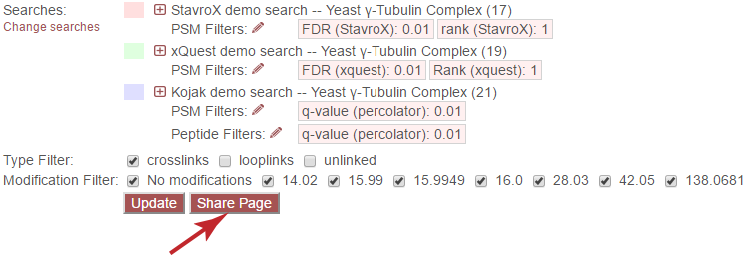
The list of merged searches is presented below the top navigation. Each search is shown next to its assigned color for the page, and the color referencing this search is retained in the Euler diagram and in the peptide table. Clicking the [+] icon will expand that search to view details:
Search Details¶
The “Path” is the location on disk from which the data were imported. The “Linker” is the name of the crossinker used in the experiment. “Search Program(s)” is the name and version number of the PSM search software used. “Upload date” is the date the data were uploaded into proxl. “FASTA file” is the name of the FASTA file used to perform the PSM search.
Search Filter¶
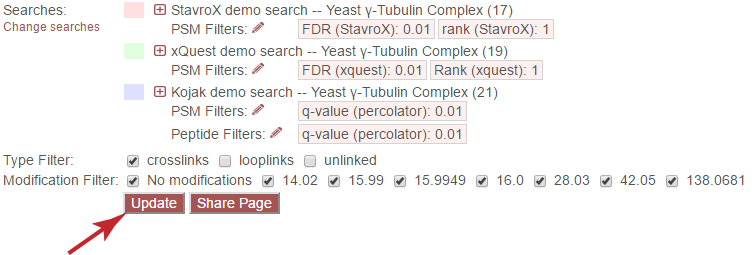
Each search is filtered separately, according to its own native score types. To change the filters for each search, click the pencil icon next to “PSM Filters:” or “Peptide Filters:” next to each search.
PSM Filters¶
The filters to apply at the PSM level. Only results which have at least one PSM that meets all of the selected critiera will be listed. When listing PSMs associated with peptides, only PSMs that meet all of the selected critiera will be listed.
To change the PSM-level filters, first click the pencil icon next to “PSM Filters”:
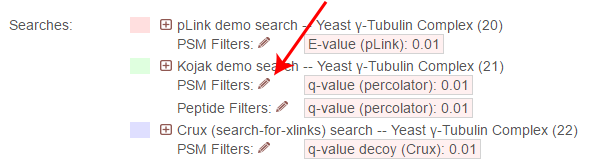
This opens an overlay with the containing the possible score types to use as PSM filters for this search. To change the cutoff values to be used for any of these score types, enter the value next to the score type. proxl will correctly handle scores for which larger values are more significant or scores for which smaller values are more signiciant.
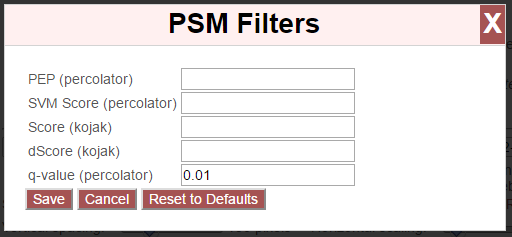
To save the new values to the page, click the “Save” button. To cancel, click “Cancel”.
The “Reset to Defaults” button will reset the cutoff values to the defaults specified by the proxl XML file uploaded to the database. This typically represents the suggested cutoffs by the author of the respective search program.
Important: It is necessary to update the data on the page after changing filter cutoff values. After clicking the “Save” button, you must click the “Update” button on the page to apply any new PSM- or peptide-level filters.
Peptide Filters¶
The filters to apply at the peptide level. Only results which have at least one peptide that meets all of the selected critiera will be listed.
To change the peptide-level filters, first click the pencil icon next to “Peptide Filters”:
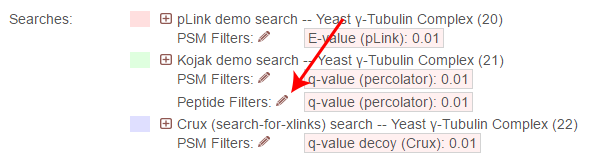
This opens an overlay with the containing the possible score types to use as peptide-level filters for this search. To change the cutoff values to be used for any of these score types, enter the value next to the score type. proxl will correctly handle scores for which larger values are more significant or scores for which smaller values are more signiciant.
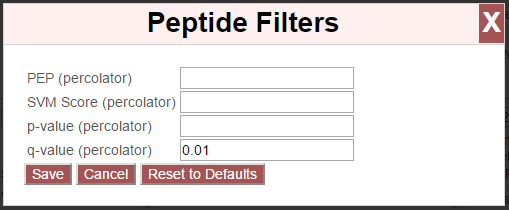
To save the new values to the page, click the “Save” button. To cancel, click “Cancel”.
The “Reset to Defaults” button will reset the cutoff values to the defaults specified by the proxl XML file uploaded to the database. This typically represents the suggested cutoffs by the author of the respective search program.
Important: It is necessary to update the data on the page after changing filter cutoff values. After clicking the “Save” button, you must click the “Update” button on the page to apply any new PSM- or peptide-level filters.

General Options¶
Change Searches¶
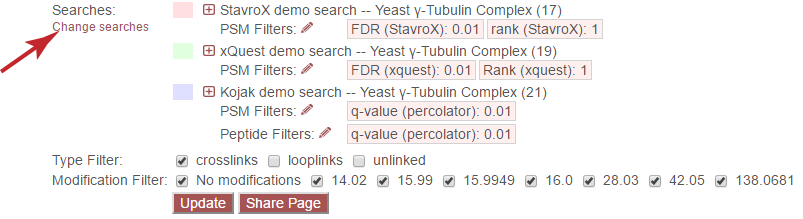
The “Change searches” link allows the user to change which searches are currently being displayed. Clicking the link causes the following overlay to be displayed:
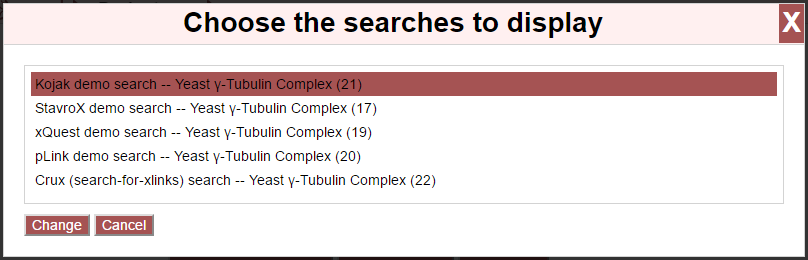
Select or de-select searches by clicking on them in the list. Once done, click “Change” to update the page with the new data or “Cancel” to close the overlay.
Update From Database¶

If the user changes any filter parameters–such as PSM/peptide score cutoffs–this button must be clicked to reflect the new filter choices.
Filter Data¶
Type filter¶
Only peptides of the checked type(s) will be returned. Proxl defines the types as:
- crosslink - A pair of peptides linked by a crosslinker.
- looplink - A single peptide with two residues linked by a crosslinker.
- unlinked - The peptide without a crosslinker on any residue.
Checking multiple boxes will include any peptide that has at least one of the checked types. I.e., checking ‘crosslinks’ and ‘looplinks’ will only include peptides that are either crosslinks or looplinks. Only checking ‘crosslinks’ will only return crosslinked peptides.
Modification filter¶
Only peptides with at least one of the checked modifications will be included. Note that monolinks are considered modifications of residues in proxl, so the mass of the crosslinker when found on monolinks is included here.
Update¶
Important: It is necessary to update the data on the page after changing filter cutoff values. After clicking the “Save” button, you must click the “Update” button on the page to apply any new PSM- or peptide-level filters.

Euler diagram¶
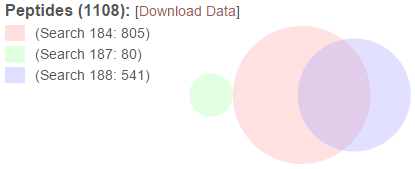
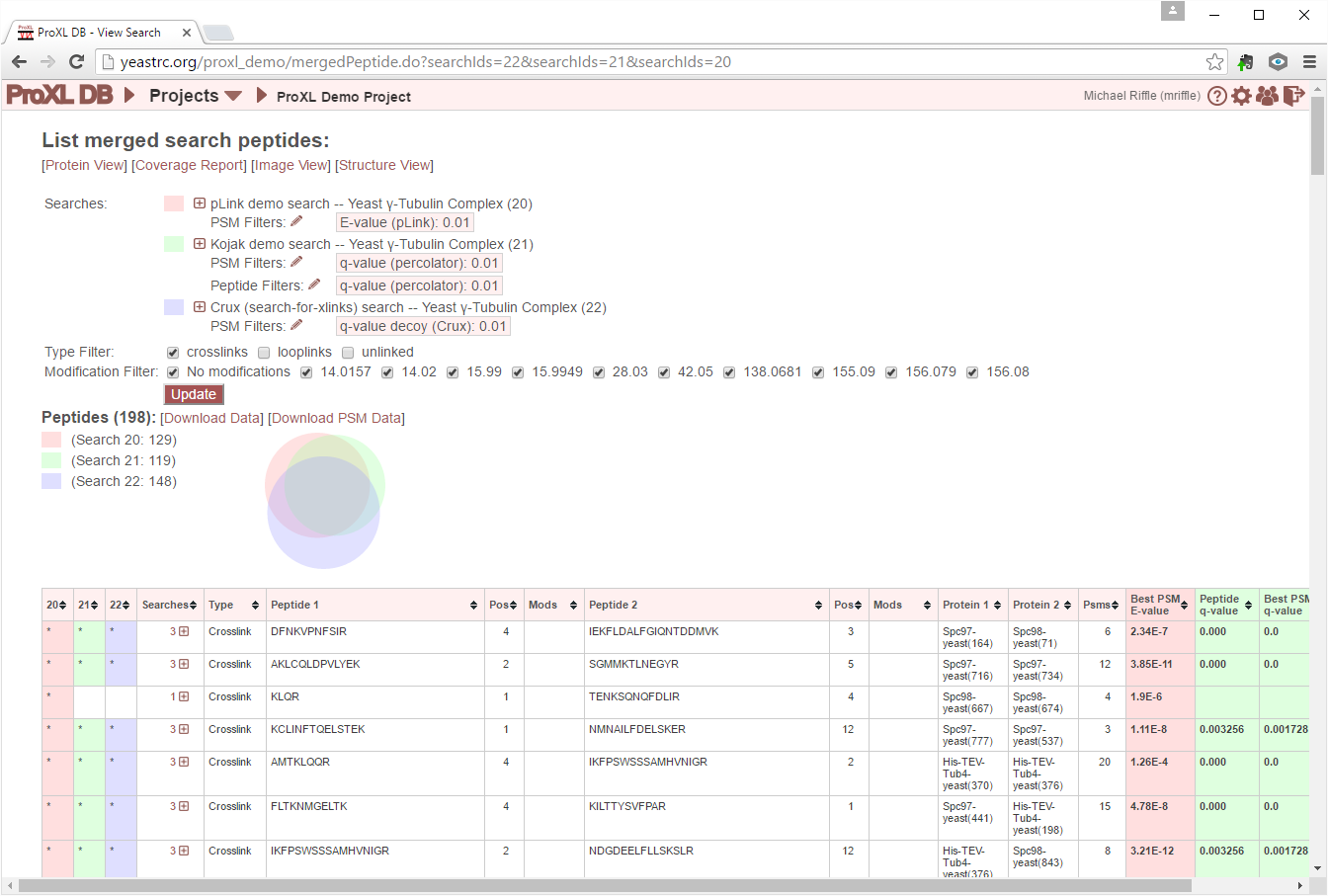
The Euler diagram (similar to a Venn diagram) provides a graphical depiction of the relative sizes and overlap between the peptides found in the merged searches. The colors in the diagram match the colors used for the search list above. The search list is provided to the left of the diagram with their associated colors as a legend. The labels for each color include the search ID number and the number of distinct peptides found in each of the merged searches. The total number of peptides resulting from the merge is presented in the header above the legend next to “Peptides”.
The “[Download Data]” link in the legend header will download the data in the table as a tab-delimited text file.
Table Description¶
The table presents columns describing the peptides and indicates in which of the merged searches the peptides were found. There is one row per peptide. A peptide on this page is defined as the unique combination of peptide sequence(s), link positions in those peptides, and dynamic modifications present on the peptides. So an unmodified peptide and a modified peptide with the same sequence will appear as separate rows in the table. Each row in the table may be clicked on to expand and view the peptide-level statistics for the given peptide from each search. Each of these searches may then be clicked on to view PSMs and spectra from those searches.
Columns¶
The columns are described below. Note that all column headers may be clicked to toggle between ascending and descending sorting of that column. Holding the shift key while clicking column headers allow sorting on multiple columns.
Search Columns¶
The first 1-3 columns will be labeled with search ID numbers as headers, and provide an indication for whether or not the peptide in that row was found in that search. If found in that search, the cell for this search in this row will be shaded the same color associated with that search in the Euler diagram and search list at the top of the page. The column will also contain an asterisk. If not found, this cell is empty.
Searches¶
The number of the merged searches that contain this peptide. The [+] icon indicates that the row may be clicked on to be expanded to show underlying searches in which this peptide as found, the stats for this peptide from each search, and the ability to view PSMs and associated spectra.
Type¶
The type of peptide (crosslink, looplink, or unlinked).
Peptide 1¶
The parsed sequence of the peptide (or the first peptide in the case of crosslinks).
Pos¶
The position in that peptide containing the linker.
Mods¶
A comma-delimited list of dynamic modifications found for peptide 1 in the form of position(mass). E.g., 17(15.99), 20(14.02)
Peptide 2¶
The parse sequence of the second peptide in the crosslink.
Pos¶
The position in that peptide containing the linker.
Mods¶
A comma-delimited list of dynamic modifications found for peptide 2 in the form of position(mass). E.g., 17(15.99), 20(14.02)
Protein 1¶
The protein(s) to which the first peptide matches, and the position in that protein to which the linker position in that peptide matched. Mouse-over the protein name to get a description.
Protein 2¶
The protein(s) to which the second peptide matches, and the position in that protein to which the linker position in that peptide matched. Mouse-over the protein name to get a description.
# PSMs¶
The total number of combined PSMs from each search for this peptide that meet the filtering critera. Note: click the table row containing the peptide to see the PSMs.
Best PSM- and Peptide-level Scores¶
Separate columns, color-coded for each search, display the best PSM- and peptide-level scores currently being used to filter the data from each search.
View PSMs¶
To view PSMs for a given peptide, first click on a row in the table to expand and view the peptide-level statistics for a given peptide from each search in which it was found (at the given cutoffs). Each of these searches may be clicked to expand and view all PSMs that meet the current filtering criteria.
Columns¶
The PSMs appear in a table with the following columns:
Scan Num.¶
The scan number from the spectral file (e.g., mzML file)
Obs. m/z¶
The observed m/z of the precursor ion.
Charge¶
The predicted charge state of the precursor ion.
RT (min)¶
The retention time in minutes.
Scan Filename¶
The filename of the scan file.
Scores¶
A column for each PSM-level score from the respective search.
View Spectra¶
The annotated mass spectrum may be viewed for any PSM by clicking the “View Spectrum” link. For help on our spectrum viewer, see the Spectrum Viewer page.
Sort Data¶
All column headers may be clicked to toggle between ascending and descending sorting of that column. Holding the shift key while clicking column headers allow sorting on multiple columns.
Download Data¶
Clicking the [Download Data] link in the header of the Euler diagram will download the shown data as a tab-delimited text file.